Performance Evaluation of ASIIQT
1. Brief Description
Like IDP-ASE [1] and HapIso [2], ASIIQT also uses the 3rd-generation Single-Molecule Sequencing (SMS) long reads to phase transcripts. HapIso only depends on SMS long reads rather than the high-throughput 2nd-Generation Sequencing (SGS) short reads. However, ASIIQT is more like IDP-ASE, both of which also use SGS reads to assist phasing. The main differences between ASIIQT and IDP-ASE include: 1) Usage of SGS and SMS reads. ASIIQT uses SGS short reads to call individual SNPs/Indels and phases isoforms mainly with SMS reads; ASIIQT also uses SGS reads to correct the error-prone SMS sequences. IDP-ASE uses SGS reads to call individual SNPs/Indels and phases isoforms with both SGS and SMS reads, and the SGS contributed much more than SMS in phasing [1]. 2) Methodology of phasing. While IDP-ASE uses elaborate statistical inference to phase the isoforms with SGS and SMS sequences, ASIIQT uses a more straightforward empirical strategy to find the linkage among SNPs from SMS long reads. 3) Correcting the SMS sequences. ASIIQT corrects the non-polymorphic bases of the SMS sequences by mapped SGS reads simultaneously. IDP-ASE does not correct the SMS long reads. The main advantage of ASIIQT is the simplicity for application, while the main drawback could be the lower sensitivity due to the no use of SGS for additional SNP phasing. The ASIIQT.V.2.0 has integrated an ‘sgsphasing’ module to add SNP pairs and isoforms phased from SGS data.
Because of the failure in implementing IDP-ASE, in this report, we only compared the performance of ASIIQT and HapIso. HapIso phases isoforms one by one and requires relatively long running time, so only 50 ASIIQT-phased transcripts were selected randomly from cDNA-normalized blood libraries to phase with HapIso. The haplotype transcripts phased by ASIIQT were also validated by SGS short-read RNA-seq (250PE, Illumina HiSeq 2000) and whole-genome sequencing (WGS) data (150x, 250PE, 400bp-insert size, PCR-free, Illumina HiSeq 2000) for peripheral blood of the same subject.
2. Performance evaluation
2.1 Comparison with HapIso
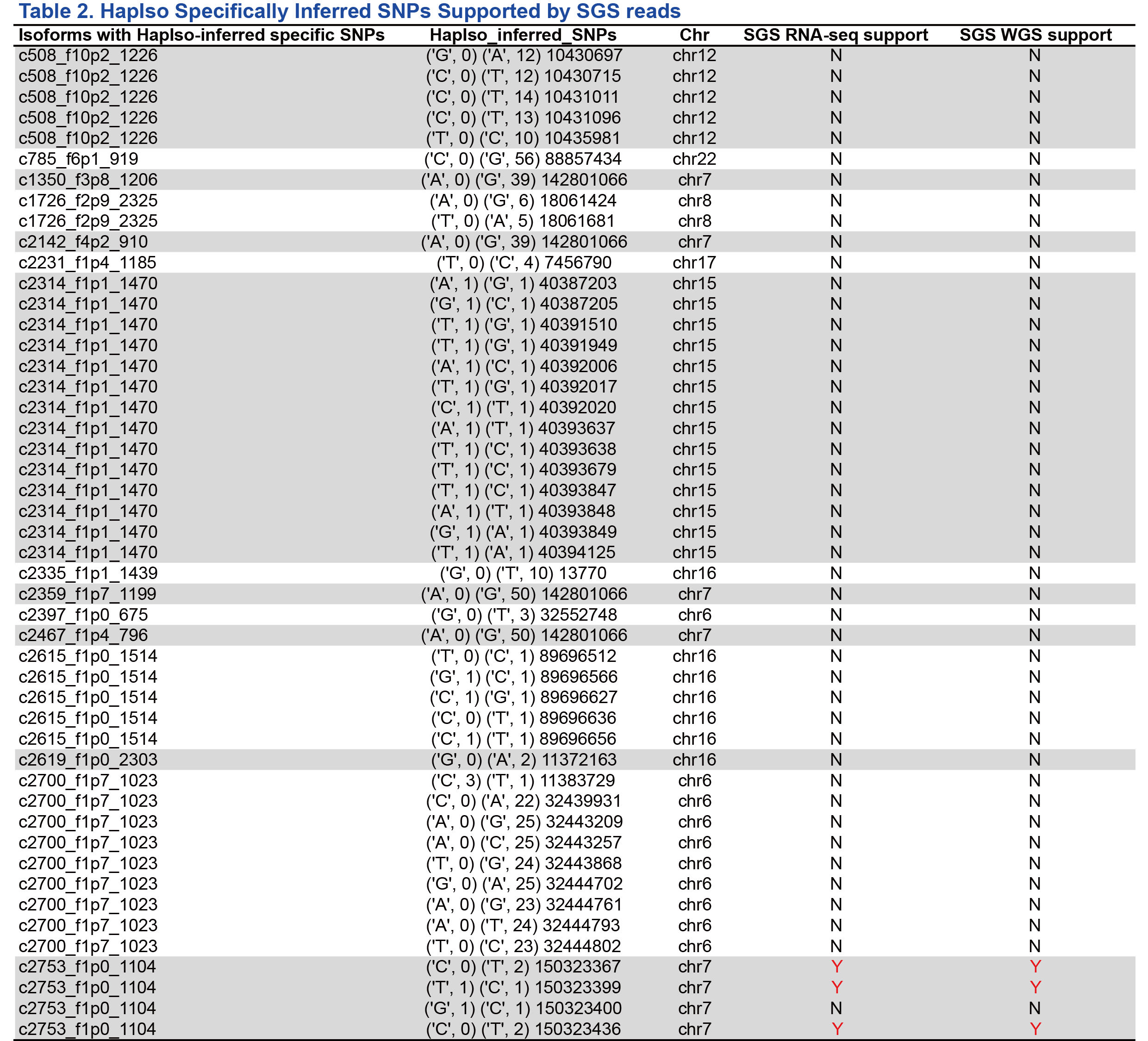
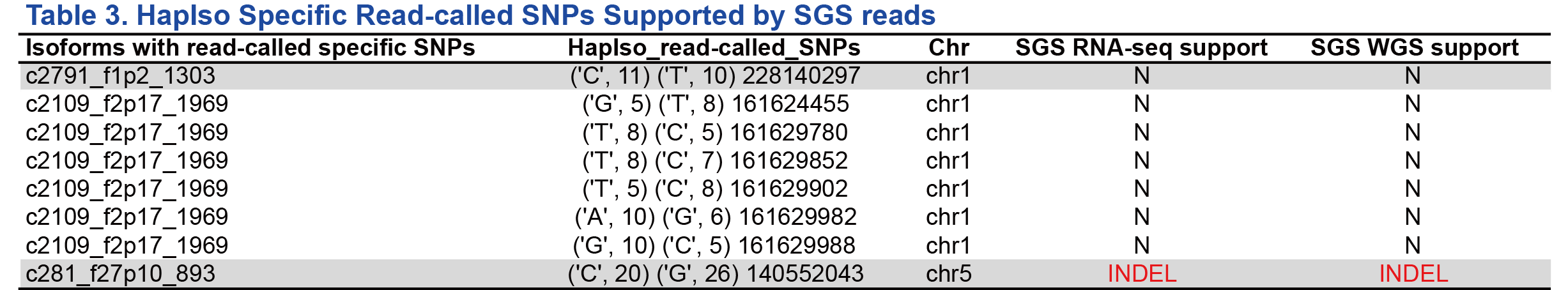
In total, 50 isoforms were selected randomly from the ones phased by ASIIQT (Table 1). Among them, only 32 (64%) could also be phased by HapIso (Fig 1A; Table 1). From the 50 isoforms, 120 and 88 SNPs were called by ASIIQT and HapIso, respectively, while 32 SNPs were phased by the two tools simultaneously (Fig 1B). A majority (48/56, 86%) of the SNPs specifically called by HapIso were inferred with no or few supported SMS reads, suggesting the relative low confidence for these SNPs (Fig 1C; Table 2). The read-called SNPs were also with low read coverage (Table 3). We further examined these SNPs from RNA-seq reads and WGS reads of the same sample. For both the read-called or inferred HapIso-specific SNPs, very few of them were called from the short reads (1/8 and 3/48 respectively, from both RNA-seq and WGS) (Fig 1D). The read-called one supported by SGS reads was in fact an INDEL, consistent with the same SNP identified and phased by ASIIQT (PB.5989.3). The three inferred SNPs supported by SGS reads were from a single isoform (PB.6868.1), which had four inferred SNPs in total but one not being supported by short reads.
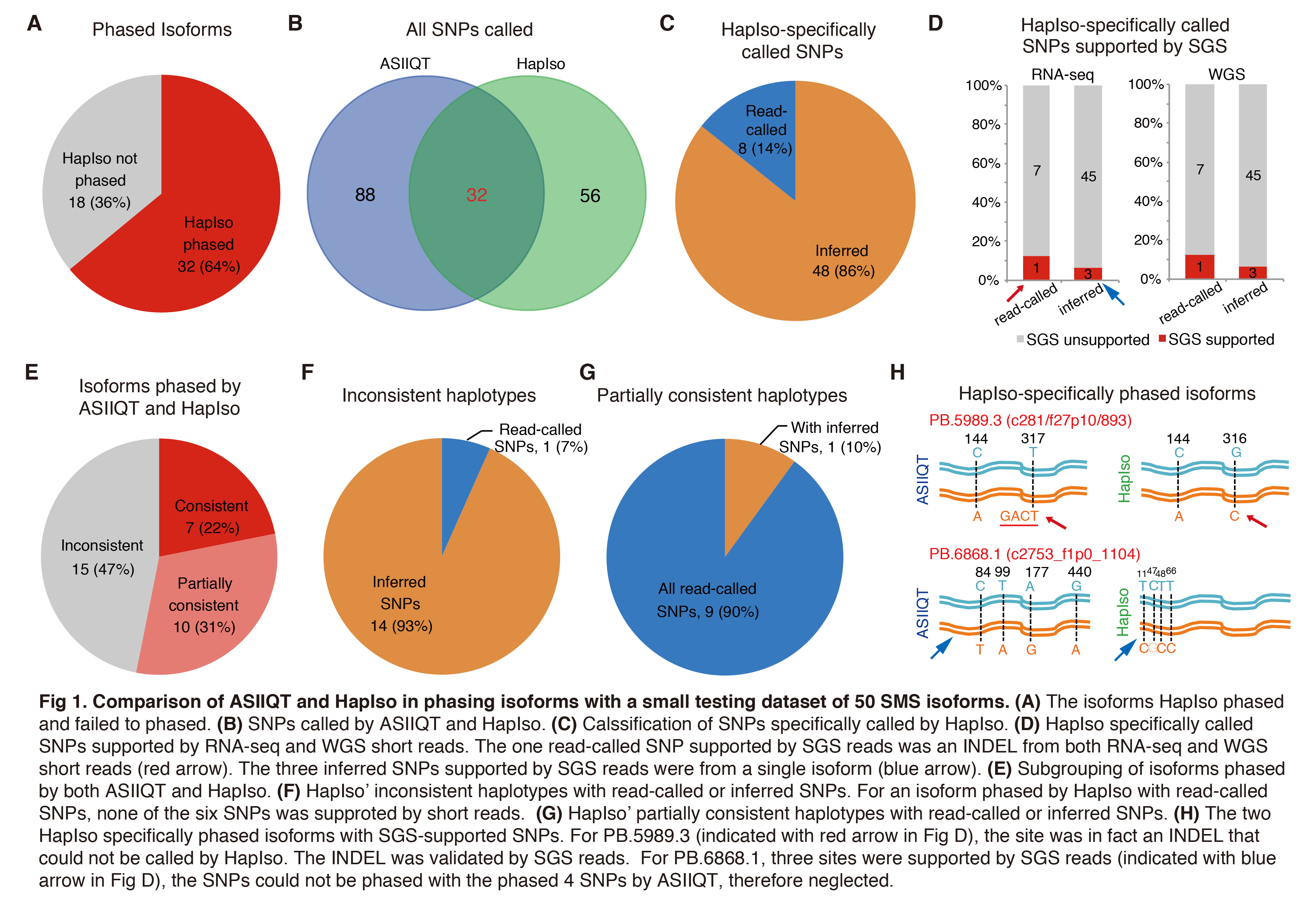
The isoforms phased by both ASIIQT and HapIso could be further divided into 3 subgroups: consistent isoforms with the SNPs phased by the two tools completely consistently (7/32, 22%), partially consistent ones with part of SNPs phased by the tools consistently (10/32, 31%), and inconsistent ones with different haplotypes (15/32, 47%) (Fig 1E; Table 1). For all the inconsistent isoforms, however, ASIIQT and HapIso generated haplotype molecules with different SNP subsets, rather than contradictory isoforms for the same SNPs (Table 1). Similarly, for the partially consistent isoforms, no contradiction was found between ASIIQT and HapIso; in contrast, HapIso often called fewer SNPs from these isoforms and discerned the same haplotypes with ASIIQT but with lower resolution. The inconsistent haplotypes of HapIso called and phased much more inferred SNPs with much lower confidence. Fourteen among the 15 (93%) HapIso inconsistent haplotypes contained inferred SNPs (Fig 1F). While for the partially consistent haplotypes of HapIso, 9/10 (90%) contained all read-called SNPs and only one (10%) had inferred SNPs (Fig 1G).
There were two isoforms with SNPs both read-called or inferred by HapIso and supported by SGS reads. PB.5989.3 was phased by ASIIQT and HapIso. For one site (144), the two tools phased it consistently; for the read-called site, however, SGS reads confirmed an INSERT while HapIso had the drawback in failure of calling INDELs and consequently called a ‘C’-SNP in the upstream site (Fig 1H, upper). Therefore, the tool tools phased different haplotype patterns. PB.6868.1 contained 4 SNP sites inferred by HapIso, which were not phased by ASIIQT (Fig 1H, lower). Three of the SNPs were supported by SGS short reads, and also called by ASIIQT. However, these SNPs could not be phased with the four phased by ASIIQT. ASIIQT selected the haplotype molecules with more phased SNPs to report, and consequently, the three SNPs were neglected though being called.
In summary, ASIIQT called more SNPs and phased more isoforms than HapIso. HapIso also showed lower accuracy for the inferred or few-read-covered SNPs and the haplotypes.
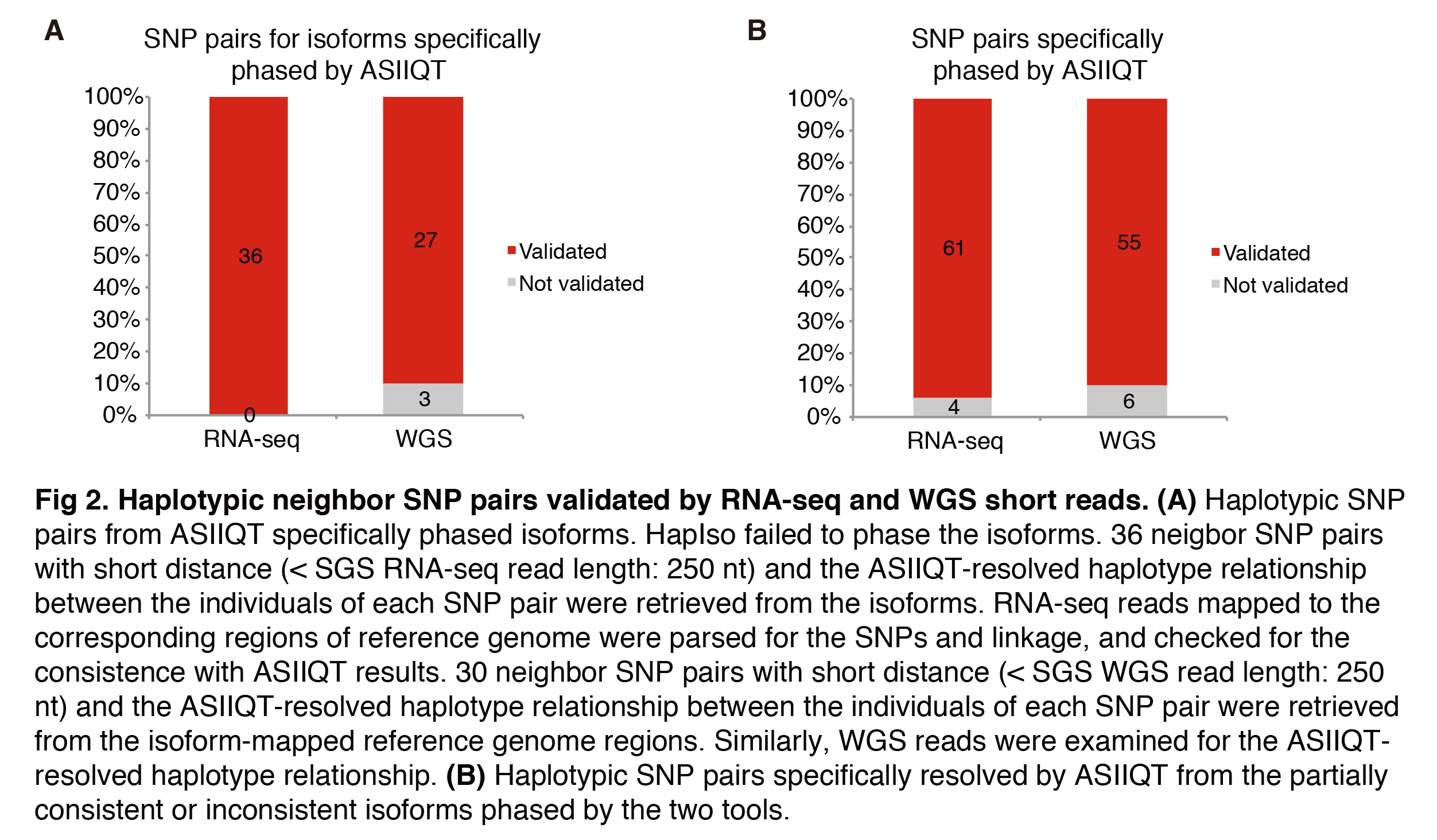
2.2 Validation with SGS reads
In total, 36 neighbor SNP pairs with short distance (<250-nt) were identified from 17 of the 18 ASIIQT specifically phased isoforms. Among them, 30 neighbor pairs (<250-nt) were identified from the genome regions of 11 ASIIQT specifically phased isoforms. These SNPs and the haplotype-linkage relationship between each pair of neighbor SNPs were examined with RNA-seq short reads and WGS short reads, respectively. All the SNPs of the 36 neighbor pairs from isoforms were recalled while the haplotype relationship between the SNPs of each neighbor pair (36/36, 100%) was found to be consistent with that phased by ASIIQT (Fig 2A). From the genome level, 27 of the 30 SNP pairs (27/30, 90%) were validated for the ASIIQT resolved haplotype relationship (Fig 2A). There were also 65 short-distance neighbor SNP pairs from transcript level for which the haplotypes were specifically phased by ASIIQT, i.e., haplotypes not phased by HapIso for the inconsistent isoforms or partially consistent isoforms. Among these pairs, 61 were identified from genome level. With RNA-seq and WGS short reads, the ASIIQT-phased haplotype relationship was validated for 61 (61/65, 93.8%) and 55 (55/61, 90.2%) pairs, respectively (Fig 2B). The non-confirmed ones were mainly due to the limited continuity of the short reads (for RNA-seq data) and the non-calling of the SNPs because of low coverage for local genomic regions (for WGS data). Taken together, the results demonstrated the high accuracy of ASIIQT in phasing isoforms and the higher sensitivity of ASIIQT than HapIso.
3. Discussion
In this report, we examined the performance of ASIIQT in phasing transcripts and compared it with that of HapIso. HapIso required more computational resources and computed the isoforms one by one, as was the reason why we selected a relatively small dataset. Because HapIso only depends on SMS long sequences to call and phase SNPs, and the raw, error-prone long reads without correction must be based on, the sensitivity and accuracy should have been influenced. We found that (1) ASIIQT could call more SNPs and phase more isoforms than HapIso, (2) the SNPs specifically identified by HapIso were often with low read coverage or inferred and were not accurate, and therefore the haplotypes for these SNPs were not accurate either, and (3) the haplotypes of most of the short-distance SNPs pairs phased by ASIIQT were validated by SGS short reads. In addition, HapIso has an inborn deficiency in calling INDELs, while ASIIQT does not have the problem since it uses SGS reads and corresponding third-party tools to call SNPs and small INDELs.
For ASIIQT, IDP-ASE and HapIso, the SNPs and their based haplotype isoforms were resolved from RNA-seq data. For exclusively mono-allele expressed isoforms, however, no SNP could be identified. Therefore, genome sequencing in parallel could further facilitate the identification of this subset of SNPs and phasing corresponding isoforms. On the other hand, the RNA transcripts often span very long regions within genome because of multiple large introns. Phased isoforms by long and short reads could also guide the assembly of haplotype genomes with longer continuity.
Reference
- Deonovic B, Wang Y, Weirather J, Wang XJ, Au KF. IDP-ASE: haplotyping and quantifying allele-specific expression at the gene and gene isoform level by hybrid sequencing. Nucleic acids research. 2017;45:e32.
- Mangul S, Yang TH, Hormozdiari F, Dainis AM, Tseng E, Ashley EA, et al. HapIso: An Accurate Method for the Haplotype-Specific Isoforms Reconstruction From Long Single-Molecule Reads. IEEE transactions on nanobioscience. 2017;16:108-115.